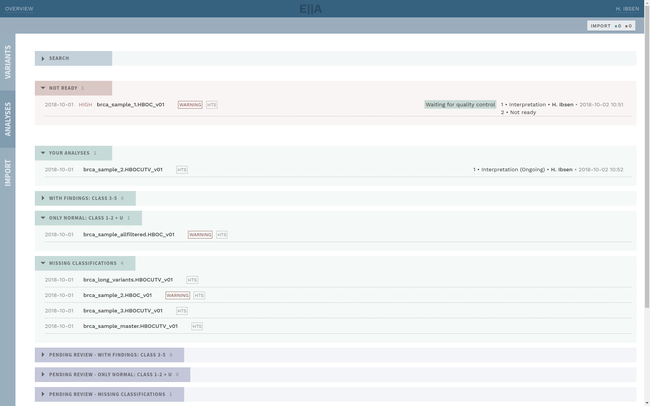
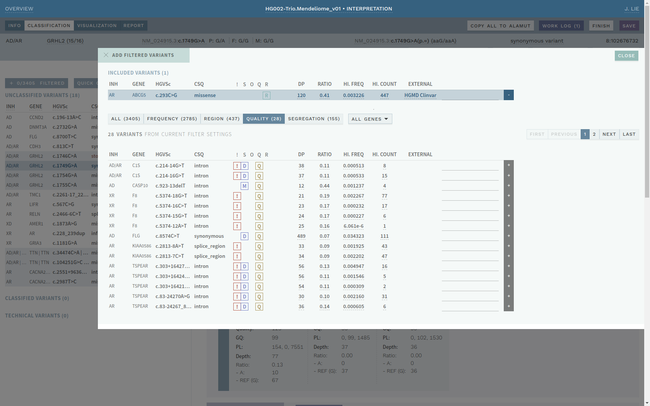
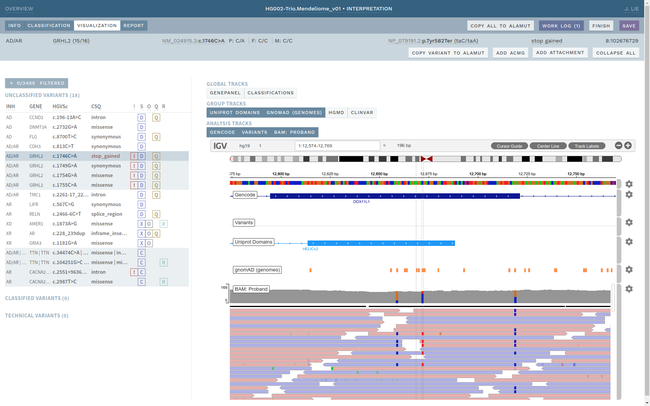
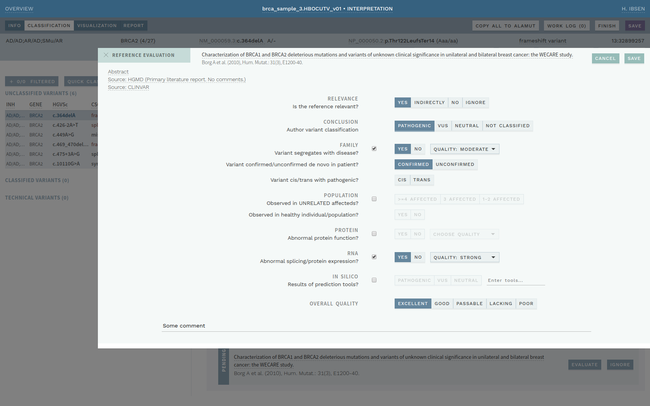
Clinical interpretation of genetic variants is a complicated process that depends on subjective judgement and unstructured evidence from research articles. This has led to variations in conclusions between laboratories, in some cases with serious medical consequences.
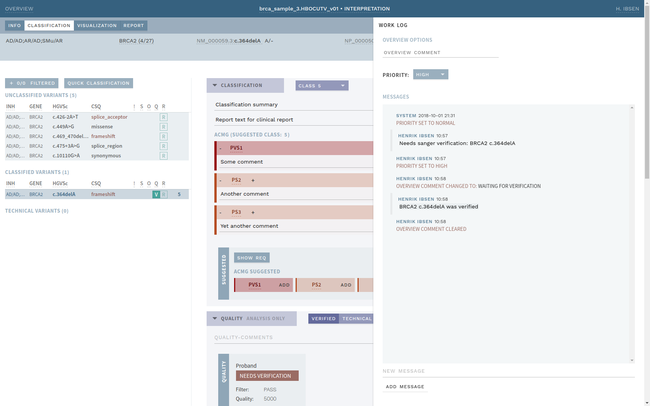
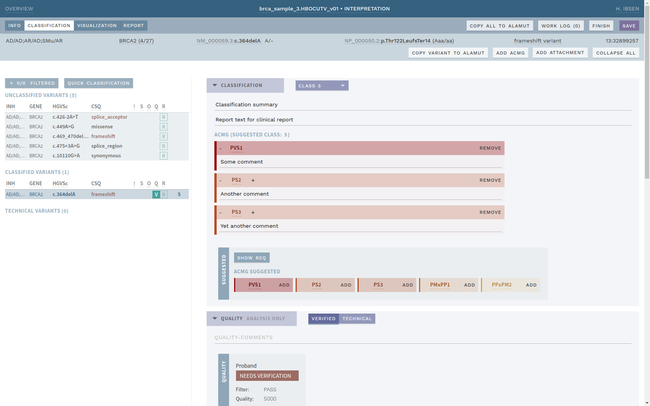
ELLA aims to reduce variability by guiding the interpreter through the various steps of an assessment based on ACMG-AMP guidelines, while simultaneously translating key facts into a format suitable for data processing. This makes it possible to automate parts of the process and suggest conclusions for the user. In addition, the justification for each assessment becomes more transparent, standardized and reproducible, which is crucial for reuse and sharing of results with other laboratories.
Information security and patient safety is at the core of ELLA, with a design that allows running in an air-gapped environment when necessary. The workflow encourages peer review of all variants of medical significance, and a complete history is kept of all changes.
ELLA works in conjunction with the annotation service anno, with post-processing and preparation of data in anno-targets (generic example or in-house). For building and maintaining clinical gene panels we also offer the Gene panel builder, which utilizes information about PanelApp panels, transcripts, inheritance modes and coverage to tailor clinical gene panels to local needs.