# Release notes: Latest releases
| Major versions | Minor versions |
|---|---|
| v1.21 | v1.21.1, v1.21.2, v1.21.3 |
| v1.20 | v1.20.1, v1.20.2 |
| v1.19 | v1.19.1 |
| v1.18 | |
| v1.17 | v1.17.1, v1.17.2, v1.17.3, v1.17.4, v1.17.5 |
| v1.16 | v1.16.1, v1.16.2, v1.16.3, v1.16.4, v1.16.5 |
| v1.15 | v1.15.1 |
| v1.14 | v1.14.1, v1.14.2 |
See older releases for earlier versions.
# Version 1.21.3
Release date: 22.05.2025
# Highlights
This is a bugfix release for two issues related to gene panels.
# All changes
- Fixed a layout bug in Gene panel info.
- Fixed an bug where custom regions from gene panels were imported as transcripts.
# Version 1.21.2
Release date: 22.04.2025
# Highlights
This is a patch release with minor changes to show custom annotation and fix layout in the Region section, improved handling of gene panels as well as minor improvements in the backend.
# All changes
- Show custom annotation in the Region section.
- Fixed truncation of long variants in the Region section.
- Custom gene panel names are now appended with time stamp in addition to date (YYMMDDTHHmm, e.g. "250422T0947") to allow for multiple imports on the same date.
- Added new CLI commands for gene panel management (activate, deactivate, and batch deactivate old versions; see
ella-cli genepanel --help). - Various code improvements and bugfixes.
# Version 1.21.1
Release date: 31.03.2025
# Highlights
This is a patch release with minor improvements in the backend.
# All changes
- Various code improvements and bugfixes
# Version 1.21
Release date: 03.03.2025
# Highlights
This release includes support for custom regions in gene panels, as well as several changes for upcoming new frontend and LIMS integration.
# All changes
- ELLA now supports custom regions in gene panels. These are exempted from any Region filter rules.
- Improved formatting for HTML tables on INFO page.
- Various improvements to new frontend (in development).
- Various code improvements and bugfixes, including adaptations for LIMS integration.
# Version 1.20.2
Release date: 17.01.2025
# Highlights
This is a bugfix release.
# All changes
- Fixed a bug in handling of reference assessments.
# Version 1.20.1
Release date: 16.01.2025
# Highlights
This is a patch release. Includes improvements to how stand-alone variants are shown and several bugfixes.
# All changes
- Opening av stand-alone variant (VARIANTS workflow) now shows the latest finalized assessment and annotation also before START, irrespective of which workflow mode was used.
- VARIANTS workflow history has been removed from variant search results to prevent confusion.
- Fixed link to documentation.
- Fixed the exception filter for generic annotation.
- Fixed a bug in finding analyses for a variant with duplicates.
- Fixed dropdown listing gene panels on import page.
- Fixed overflows for variant history with large images.
- Improved efficiency of the generic annotation filter.
- Various code improvements and bugfixes.
# Version 1.20
Release date: 03.09.2024
# Highlights
This release provides the possibility to use any available annotation in filtering rules and numerous updates and improvements to the code base.
# All changes
- ELLA can now be configured to use any available annotation in filtering rules. See Genereic annotation filter for details.
- Dropdown lists of gene panels (on import page or when viewing standalone variants) now only shows latest version of each gene panel, correctly sorted.
- It is now possible to create templates for comment field in the Region section (Classification page).
- Fixed a bug causing empty variant tracks for analyses with both SNV and CNV.
- Webhooks on selected endpoints are now supported. See Webhooks for details.
- New frontend (in development) merged into repo from ella-frontend (opens new window). See Development and Testing for details.
- Various code improvements and bugfixes.
# Version 1.19.1
Release date: 17.06.2024
# Highlights
This is a patch release.
# All changes
- Code improvements and bugfixes.
# Version 1.19
Release date: 22.03.2024
# Highlights
This release provides no new or changed features, but brings numerous updates and improvements to the code base.
# 🔺 Breaking changes for 1.19
This release brings several significant changes to deployment. See Production for details.
# All changes
- Upgrades and improvements to code base and development environment, including use of Docker Compose instead of a Makefile and upgrade to Python 3.12 (with corresponding features utilized).
# Version 1.18
Release date: 08.01.2024
# Highlights
This release provides no new or changed features, but brings numerous updates and improvements to the code base and significant performance enhancements.
Note the possibility for breaking changes for this version.
# 🔺 Breaking changes for 1.18
# Potential problems with database migration
This release includes an upgrade of Alembic, which might break database migration (ella-cli database upgrade head) from versions earlier than v1.17.2 to versions v1.18 and later. If you experience problems, perform the migrations in two steps:
- Run migrations using any version v1.17.2-v1.17-5.
- Run migrations using latest/desired version (v1.18.0 or later).
# All changes
- Upgrades and improvements to code base: Includes upgrades to Python packages, PostgreSQL (now at v15) and SQLAlchemy (now at v2.0.21) and use of Poetry instead of Pipenv. Changes also result in significantly improved performance of database queries.
- Fix bug in ACMG suggestions for reference evaluations.
# Version 1.17.5
Release date: 29.08.2023
# Highlights
This is a patch release.
# All changes
- Show analysis specific quality information for Sanger variants. Allows custom FILTER tags on the VCF to trigger warnings.
- Fix bug causing unavailability of Worklog templates.
# Version 1.17.4
Release date: 28.06.2023
# Highlights
This is a bugfix release.
# All changes
- Allow de novo probability computation to fail (circumvents a rare bug where an analysis containing an X-linked variant fails to load due to a mismatch between gender and computed genotype probabilities).
# Version 1.17.3
Release date: 26.06.2023
# Highlights
This release adds a small modification to the ACMG rules engine for Suggested class and several UI improvements and bugfixes.
# All changes
- ELLA now suggests Class 5 (not 4) for combination 1 PVS + >=1 PM, based on recommendations from Tavtigian (2018), ACGS (Ellard 2020) and CanVIG.
- "Worse consequence" warning is now shown also for CNVs.
- Improved display of information in top bar, popovers, and Consequence and Quality cards.
- Allow annotation with empty transcript
SOURCE, e.g. if using VEP--refseqoption. - Fixed a bug causing error on loading ELLA for users with read-only access.
- Fixed a bug causing non-persistance of chosen user groups when importing using custom gene panels.
- Fixed a bug causing manual imports to fail when multiple chosen user groups have the same config.
- Fixed a bug causing the Report page to crash when there are many transcripts for an included variant.
- Improved help text for variant import.
- Changed license from MIT to GNU GPL v3.
# Version 1.17.2
Release date: 09.05.2023
# Highlights
This release fixes a bug and adds support for non-coding transcripts and showing PL value in the Quality card.
# All changes
- Add support for non-coding transcripts.
- Add Phred-scaled genotype likelihood (PL) (opens new window) value to the Quality card.
- Fixed a bug causing errors when inspecting the work log for a variant interpretation with no allele report.
# Version 1.17.1
Release date: 26.04.2023
# Highlights
This is a bugfix release.
# All changes
- Fixed a bug causing inability to finalize previously evaluated allele assessments.
- Fixed a bug in parser and fetcher for PubMed records.
- Fixes and improvements to development environment and code base.
# Version 1.17
Release date: 28.03.2023
# Highlights
This version does not add significant new features, but switches to a new, improved format of gene panels.
Note there are several breaking changes with this version, and admins should take care to update the necessary files.
# Improved format of gene panels
The biggest change in this release is a new setup for gene panels. Although this doesn't add any new features in ELLA by itself, the new process for building, updating and configuring gene panels represent a significant improvement, both in terms of more easily keeping transcript and gene names updated and in terms of keeping manual configuration consistent across gene panels.
For details, see the Gene panel builder (opens new window) documentation and the separate GitLab (opens new window) repository.
Note that these changes also make providing gene panel version in the usergroup config optional; see User groups and gene panels and Default import gene panel.
# 🔺 Breaking changes for 1.17
# Gene panels must be updated for new imports
Changes in this version deprecates all gene panels in the old format. Historic analyses imported with old panels will still load, but no new imports will be possible until gene panels have been updated to the new version. See above for details.
# Changes to user configuration
The configuration options allow_notrelevant and allow_technical have been removed. To update:
Remove
allow_notrelevantandallow_technical(if any) from your user group config (usuallyfixtures/usegroups.json).Remove
allow_notrelevantandallow_technicalfrom your application config (underuser.user_config.workflows.analysis.finalize_requirementsinella_config.ymlor similar; see UI options).Update user groups with:
ella-cli users add_groups <path to usergroups.json>
# Removal of export scripts
The CLI command ella-cli export classifications has been removed and export scripts will no longer be maintained as part of this source code. The last version of the script is available here (opens new window).
# All changes
- Improved format of gene panels.
- Added link to UCSC Genome Browser (opens new window) in chromosome position in the top bar.
- Upgraded IGV.js in VISUAL to v2.12.6 (opens new window). This includes fixes for coloring items by pair orientation and insert size and bugs that affected rendering of bed files and amino acid translation.
- Added support for bigBed file format in VISUAL.
- Added support for VEP v108.2 (opens new window).
- Removed config option for disallowing Technical and Not relevant for Finalize (set to always allowed).
- Removed export scripts.
- Upgraded Python to v3.11 (opens new window) and pydantic to v1.10.5 (opens new window).
- Updated parser and fetcher for PubMed records.
- Fixed a bug causing gene assessments not being loaded for variants where default transcript is missing from annotation.
- Fixes and improvements to development environment and code base.
# Version 1.16.5
Release date: 30.01.2023
# Highlights
This is a bugfix release.
# All changes
- Refined some queries that used sequential scans on large tables due to how they were formed
# Version 1.16.4
Release date: 08.03.2022
# Highlights
This is a bugfix release.
# All changes
- Upgraded IGV.js in VISUAL to v2.11.0 (opens new window), using a temporary fork (opens new window) with additional bugfixes that remove excessive error messages in VISUAL.
# Version 1.16.3
Release date: 02.03.2022
# Highlights
This is a minor release with a few bugfixes, but also includes an upgraded IGV and track descriptions available as a mouseover on the track selection buttons in VISUAL.
# All changes
- Added track description in mouseover for track selection buttons in VISUAL.
- Upgraded IGV.js in VISUAL to v2.10.5 (opens new window). This includes some bugfixes and adds the option "colour alignments by insert size and pair orientation".
- Added support for bgzipped VCF files.
- Fixed a bug causing a reset of analysis names when clicking import options for batch imports.
- Fixed a bug causing analysis to fail loading when PL values for de novo calculation is very high.
- Fixes and improvements to development environment and code base.
# Version 1.16.2
Release date: 21.12.2021
# Highlights
This is a bugfix release.
# All changes
- Fixed a bug where genotype display is not shown in allele bar
# Version 1.16.1
Release date: 16.12.2021
# Highlights
This is a bugfix release.
# All changes
- Fixed a bug causing inability to paste plain text into comment fields.
- Fixed word wrapping in comment fields.
# Version 1.16
Release date: 16.12.2021
# Highlights
# Experimental support for CNV interpretation
This version introduces experimental support for copy number variants (CNVs).
When enabled, a separate CNV mode is added (switched to using a button in the top bar), where CNVs are shown in a separate variant list and allow documenting, reporting and reusing CNV interpretations, as for SNVs. The current version supports CNV deletions, duplications and tandem duplications (VCF format: DEL, DUP and DUP:TANDEM, respectively; for variants of type = SVTYPE).
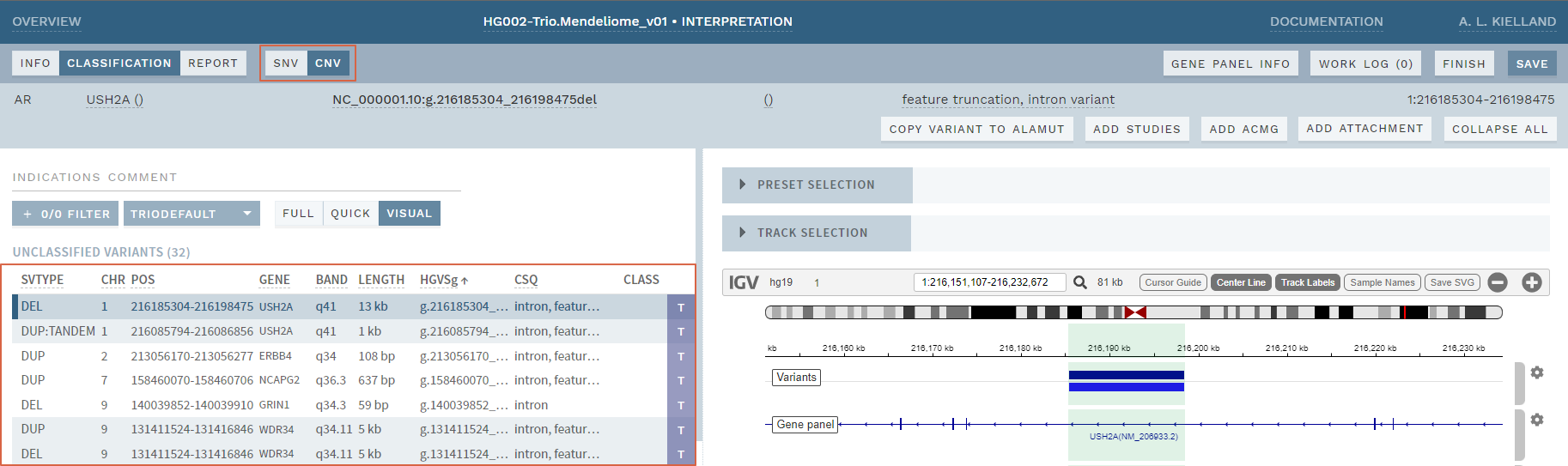
Figure: Experimental CNV mode. Major changes from SNV mode are highlighted with red squares.
Note the limitations in this version:
- No CNV filtering is done in ELLA, for the time being this needs to be done upstream, prior to import.
- No particular adaptations have been made to presentation of annotation. However, any annotation from the VCF can be added in the configuration, and we recommend adding CNV-specific tracks to VISUAL to aid in interpretation.
- No CNV-specific ACMG criteria have been added.
- No manual import of CNVs is supported.
DISCLAIMER
As the CNV features are not fully validated, this functionality is currently disabled by default, and we recommend not enabling it in production. For testing, the new features can be enabled by setting the feature flag ENABLE_CNV to true in the application configuration.
# Changes in VISUAL
Configuration of tracks in VISUAL has been further improved, allowing all types of tracks to be placed under presets. To allow better zoom/scroll control with many tracks visible, this version also adds a requirement to hold the Shift or Alt key when zooming with the mousewheel.
# 🔺 Breaking changes for 1.16
# Configuration of tracks in IGV
With this version, configuration of tracks shown in IGV needs to be updated.
Specifically, all tracks are now configured in a single config file and specified as either DYNAMIC, STATIC or ANALYSIS tracks. This means all tracks (including dynamic and analysis tracks) are now configurable, but also means that the individual track JSON config files used in earlier versions will no longer work.
To configure a particular track, the file path must be matched with a regular expression (regex). Note that the JSON format requires special regex characters to be double-escaped (e.g. .bed.gz should be written as \\.bed\\.gz). See track_config_default.json (opens new window) for examples and IGV in VISUAL for further details.
# All changes
- Added experimental support for CNV interpretation.
- Improved configuration of tracks in VISUAL, including dynamic and analysis tracks.
- Mousewheel zoom in VISUAL now requires holding the
ShiftorAltkey. - Added default transcript to Gene info.
- Updated order of VEP consequences to match the Ensembl default (opens new window).
- Fixed another bug in listing "5 most similar gene panels" for analyses with custom or older gene panels.
- Fixes and improvements to performance, development environment and code base.
# Version 1.15.1
Release date: 10.11.2021
# Highlights
This is a bugfix release.
# All changes
- Fixed a bug causing missing transcripts in exported classifications.
- Fixed a bug causing an error when listing "5 most similar gene panels" for analyses with custom gene panels.
- Fixed a bug causing IGV search to occasionally return wrong gene when there are partial matches.
- Fixed a bug causing invisible INH values for filtered variants.
- Upgraded IGV.js in VISUAL to v2.10.4 (opens new window).
- Fixed a bug causing error on start if a Worklog comment was just added.
# Version 1.15
Release date: 13.10.2021
# Highlights
This update brings a number of enhancements to the user interface, with further improvements to VISUAL, new quick add actions, as well as several smaller improvements and bugfixes.
# Improvements to VISUAL
Selecting tracks in VISUAL has been further improved by splitting off presets in one section and the individual tracks in another, with the latter section collapsed by default. Tracks are now also organized under headers determined by preset names instead of the previous "Global", "Group" and "Analysis" headers:
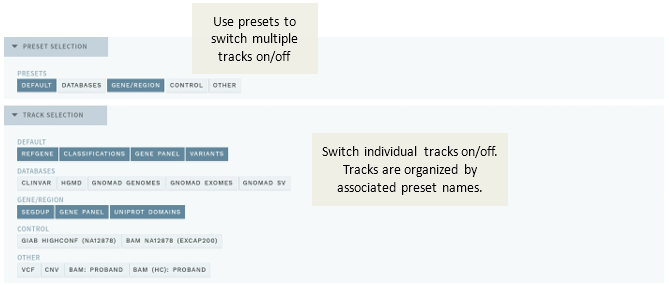
Figure: Improved track selection presets, with tracks organized by preset name.
Note that tracks that have no configuration (e.g. at present, analysis raw data) are placed in the preset OTHER .
In addition, IGV.js was upgraded to the most recent version (opens new window), which adds exon numbering (in feature popup) to transcript reference tracks, and improved performance. It is now also possible to open analyses with no unfiltered variants, e.g. for reviewing raw data before reporting negative test results.
See Visual for further details.
# New quick add actions
It is now possible to quickly add a signature and date ([username, yyyy-mm-dd]) to any comment field, using either the keyboard shortcut Alt + S or a button on the formatting toolbar:
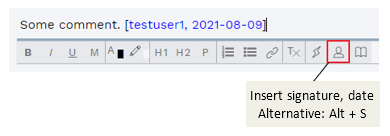
Figure: Quick add signature to any comment field.
In addition, the option to quickly add a reference was added to the Report and Indications comment fields on the REPORT page, as well as in the Indications comment in the sidebar:
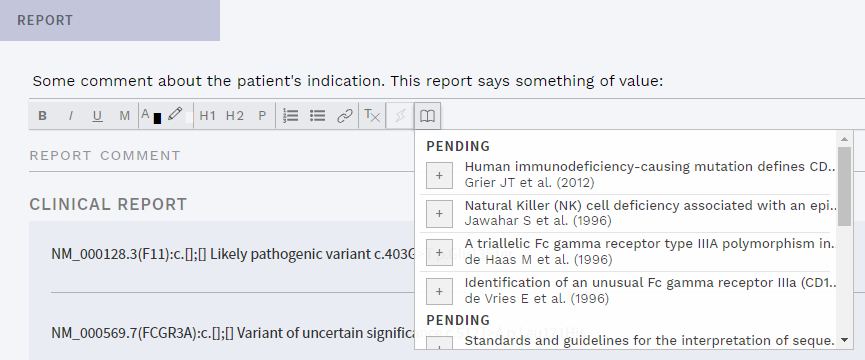
Figure: Quick add references to Indications or Report comments.
Note that for these particular comment fields, only references from variants that have been included in the REPORT are listed.
# Miscellaneous improvements to the user interface
Other user interface changes in this version include a better collapsed mode for sections on the CLASSIFICATION page, more concise gene and gene panel related information, improvements to layout, and various bugfixes.
# All changes
- Improved track selection in VISUAL with grouping of tracks under preset names.
- Upgraded IGV.js in VISUAL from v2.7.9 to v2.10.0 (opens new window) (using temporary fork (opens new window) with bug fix).
- In VISUAL, tracks with no configured preset are placed in an
OTHERpreset. - Opening VISUAL is now possible also for analyses with no unfiltered variants.
- Added button and keyboard shortcut for quick insertion of signature and date in comment fields.
- References from included variants can now be inserted in Indication and Report comments.
- Added
SHOW ANALYSESbutton to VARIANTS workflow. - Collapsing sections on the CLASSIFICATION page now also truncates comments, improving overview when comments are lengthy.
- Layout in Studies & references section is improved, removing some headers and adding an
UNPUBLISHEDtag when relevant. - Gene
INFOtag in top bar is now removed if gene comment is emptied. GENE PANEL INFO(button in top bar) now only lists latest version of each gene panel in "5 most similar gene panels".- User dashboard now only shows official (not custom) gene panels.
- Fixed a bug causing external links for currently selected variant instead of filtered variant in Filtered variants modal.
- Fixed a bug causing incorrect sorting of unpublished studies.
- Fixed a bug where variants in pseudo-autosomal regions threw errors on loading gnomAD hemizygous counts.
- Fixes and improvements to development environment and code base.
# Version 1.14.2
Release date: 31.08.2021
# Highlights
This release adds a few bugfixes.
# All changes
- Fixed a bug causing inability to import VCFs with no variants.
- Fixed missing tooltip for reference title.
- Fixed a bug causing incorrect sorting of variants in the Region section.
- Fixed a bug causing display of info from currently selected variant instead of filtered variant in Filtered variants modal.
- Fixed a bug causing inability to manually add External or Prediction info in certain instances.
- Fixed a bug causing missing hemizygous counts for legacy/default frequency annotation.
# Version 1.14.1
Release date: 08.07.2021
# Highlights
This release adds a single bugfix.
# All changes
- Fixed a bug causing front end to break in some edge cases.
# Version 1.14
Release date: 30.06.2021
# Highlights
The most significant change in this release is the addition of support for configurable annotation. In addition, several improvements have been made in the UI in preparation for CNV support.
# Support for configurable annotation
Adding new kinds of variant annotation in ELLA has up until now required changes to the source code, and has been a major limitation in the software. Starting with this release, however, new annotation can be added with a few changes to configuration. This allows much more flexibility and ease when adding new variant annotation resources. See the technical docs for more information on how to use the new configuration.
Unless new annotation is added, no changes will be visible to the end user, except a very minor change in the sorting of ClinVar entries (now sorted on date only).
# New REGION section
A new section termed REGION has been added to the CLASSIFICATION page. This shows previously classified SNVs from the internal database VarDB that are within a preconfigured genomic distance from the currently selected variant:
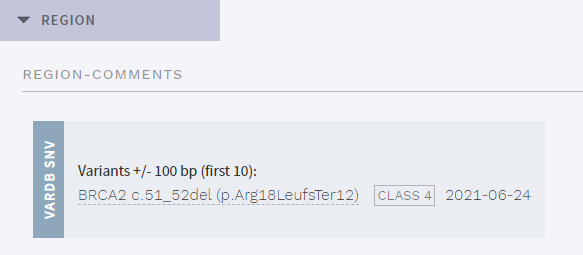
Figure: New section with nearby classified variants.
# Improvements to VISUAL
This version adds several improvements to how the VISUAL mode (with IGV.js) works. Most significantly, the track selection section on top of the VISUAL page is now collapsible and has the possibility for adding presets that allow quick selection/deselection of groups of tracks:
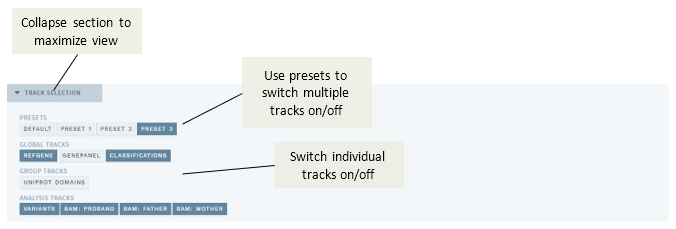
Figure: Improved track selection with possibility for presets.
In addition, the Classification track now includes links to existing allele assessments. Click a variant in the track, then the link in the resulting popover to go to the variant:
Figure: Classification track now has links to existing allele assessments.
Lastly, it is now possible to zoom the view quickly using the mouse wheel, and clicking a selected variant in the side bar recenters the view on the variant.
# 🔺 Breaking changes for 1.14
The following changes must be made to ella-config.yml to use this version:
- Remove
frequencies.viewand instead add to the newannotation-config.yml(see Annotation). - Add
similar_alleleswith subkeysmax_variantsandmax_genomic_distance(see Region).
# All changes
- Added support for configurable annotation.
- Added new section REGION on CLASSIFICATION page, showing nearby SNV assessments.
- Added support for track selection presets in VISUAL.
- Made track selection section in VISUAL collapsible.
- Enabled links to existing classifications in VISUAL.
- Added mouse wheel zoom and possibility to recenter on selected variant in VISUAL.
- Added support for
bigWigandcramtrack file formats in VISUAL. - Fixed a bug causing inability to update REPORT.
- Fixes and improvements to development environment and code base.