# Release notes: Older releases
| Major versions | Minor versions |
|---|---|
| v1.13 | v1.13.1, v1.13.2 |
| v1.12 | v1.12.1, v1.12.2, v1.12.3 |
| v1.11 | v1.11.1, v1.11.2, v1.11.3 |
| v1.10 | v1.10.1 |
| v1.9 | v1.9.1, v1.9.2 |
| v1.8 | v1.8.1, v1.8.2 |
| v1.7 | |
| v1.6 | v1.6.1, v1.6.2 |
| v1.4 | v1.4.1 |
| v1.3 | v1.3.1 |
| v1.2 | |
| v1.1 | v1.1.1, v1.1.2 |
# Version 1.13.2
Release date: 19.05.2021
# Highlights
This release changes thresholds for verification warnings and adds a few other tweaks and bugfixes.
# All changes
- Thresholds for the "Needs verification" warning was adjusted to depth <20 (was ≤20) and allele ratio (heterozygous) ≤0.3 or ≥0.7 (was ≥0.6).
- Disallow spaces and underscores in custom gene panel names when ordering reanalyses in the IMPORT module.
- Fixed a bug causing de novo likelihood calculation to fail in certain instances.
- Fixed a bug causing missing source information for studies and references.
# Version 1.13.1
Release date: 16.04.2021
# Highlights
This release adds a single bugfix.
# All changes
- Fixed a bug causing excessive load on backend.
# Version 1.13
Release date: 09.04.2021
# Highlights
This release brings several improvements to variant filtering rules, as well as a number of smaller fixes.
# Improvements to variant filters in ELLA
It is now possible to configure the Classification filter to only consider classifications that are still valid. With this option enabled it is possible to define that e.g. class 1 and class 2 variants should be filtered only if they have a classification that is still valid (not outdated).
# Improvements to pre-filters
The pre-filters (applied before import of variants into ELLA) are now configurable and has the added option of pre-filtering variants with low mapping quality (MQ<20). This latter option is relevant e.g. for variants called with Dragen-GATK, which unlike GATK does not automatically exclude variants with a low MQ.
# Upgraded IGV in VISUAL
IGV.js on the VISUAL page has been upgraded to v2.7.9 (opens new window). For ELLA users, this fixes a few bugs, but also brings new view mode options: Click the cog wheel to the right of a track to switch between "expand" (default), "squish" or "collapse" display modes (available options depend on track type).
# 🔺 Breaking changes
With the improvements to pre-filters, the configuration in usergroups.json must be updated. The equivalent to the previous
"prefilter" = True is now "prefilter": [["hi_frequency", "no_nearby_variant", "no_classification", "not_multiallelic"]]. See pre-filters for further details.
# All changes
- Added possibility for excluding outdated classifications in the Classification filter.
- Added configurability and options for pre-filters.
IGV.jshas been upgraded to v2.7.9.- Tweaked front-end error reporting to reduce number of "An error occurred ..." messages displayed to users.
- Replaced custom vcf parser with cyvcf2 (opens new window).
- Added blacklist option in the analysis watcher, allowing exclusion of specific analyses during automated import.
- Several fixes and improvements to development environment and code base.
# Version 1.12.3
Release date: 19.02.2021
# Highlights
This release adds a few bugfixes.
# All changes
- Fixed a bug causing loading of certain historical analyses to fail.
- Fixes to deposit and backend.
# Version 1.12.2
Release date: 29.01.2021
# Highlights
This release adds a few bugfixes.
# All changes
- Fixed a bug causing ELLA not to start.
- Fixed a bug causing excessive memory use in exports of variant interpretation database.
- Fixed an error in test configuration.
# Version 1.12.1
Release date: 19.01.2021
# Highlights
This release adds a few bugfixes.
# All changes
- Fixed a bug that caused pre- and postprocessing to fail.
- Fixed a bug that caused missing "Requested" date on imported analyses.
- Fixed a bug that caused performance problems.
# Version 1.12
Release date: 13.01.2021
# Highlights
This release includes many fixes and improvements, both to the frontend and backend/development environment. The most significant changes for users include changes to classification, history and variant warnings:
# Redefined classification choices
Variant classification choices have been redefined in line with ClinVar definitions (opens new window):
CLASS Uwas renamed toNOT PROVIDED, meant for recording various information (literature/research/clinical/phenotyping) without interpreting clinical significance. It is recommended to configure this class to be immediately outdated.- The choice
RISK FACTORwas added, meant for variants that are interpreted not to cause a disorder but to increase the risk.
The classification choices are now:
Figure: Redefined variant classification choices.
# Improvements to history
History for changes to the variant CLASSIFICATION REPORT field was added, and the HISTORY pop-up now shows a drop-down for viewing previous versions instead of listing them:
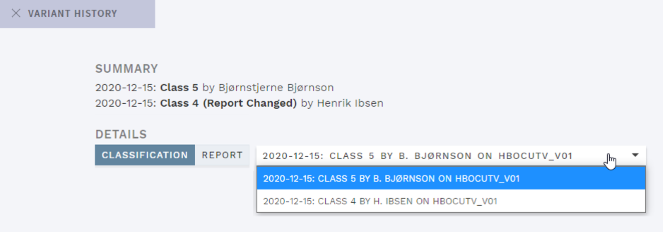
Figure: Modified variant history view with addition of Report history and drop-down for previous versions.
In addition, when opening a previously finalized analysis, ELLA will now default to displaying the state corresponding to the latest interpretation round, i.e. showing variant interpretations, classifications and annotation exactly as they were presented at the time of the last finalization. The previous default, CURRENT DATA (along with any other interpretation round), is still available using the drop-down in the top bar. To prevent confusion, a warning was added when viewing historical data.
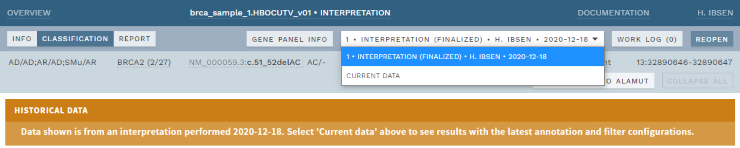
Figure: Latest interpretation round now selected by default (with warning) for historic analyses.
# Improved warnings
To improve visibility of the different variant warnings displayed on the CLASSIFICATION page, collision warnings are now shown in a yellow banner separate from annotation and user group warnings (red), and collision warnings are no longer included in the ! tag in the sidebar:
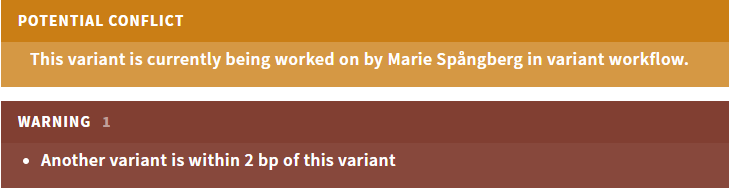
Figure: Collision and annotation warnings are now separate.
In addition, ELLA now displays a message at the bottom of the page if a finalized variant in an ongoing analysis has been updated and finalized by another user:
Figure: Message when evaluation has been updated by another user.
Note that the message is only displayed when a user finalizes another variant or manually refreshes the view.
# Select user group on custom imports
To better support custom gene panel imports that should be available to more than one user group, the custom gene panel import dialogue now includes an option to select user groups:
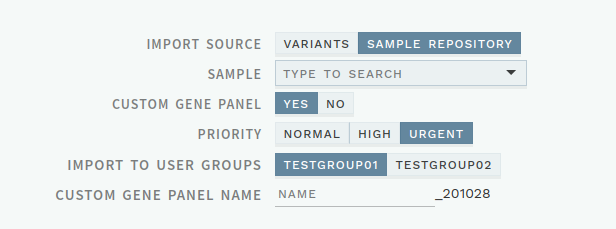
Figure: Message when evaluation has been updated by another user.
# All changes
- Redefined classification choices in line with ClinVar definitions.
- Improved design of the history modal, with addition of CLASSIFICATION REPORT history and sorted ACMG criteria.
- Show latest interpretation round by default for historical analyses, including warning.
- Variant collision warnings are now separated from annotation warnings on the CLASSIFICATION page.
- Display message when variant in ongoing analysis has been updated by another user.
- Added possibility to select which user groups an imported custom gene panel analysis should be available to.
- Search result limit has been increased from 10 to 100, with pagination of the search results.
- Added more external links in the gene information popup (ClinGen, PanelApp and ACMG incidental findings).
- Updated HGMD Pro links to point to new base url.
- Added support for templates in the Gene information comment field.
- Added possibility for linking attachments on the INFO page.
- Made allele list in top bar scrollable when number of transcripts exceed 3.
- Fixed wrong tooltip on
SUBMIT REPORTbutton. - Fixed a bug that caused variants that were removed from the REPORT to be added back again without user intent.
- Fixed a bug that caused errors in loading of historical data.
- Many fixes and improvements to development environment and code base.
# Version 1.11.3
Release date 11.11.2020
# Highlights
This release adds a bugfix for the frequency filter.
# All changes
- Fixed a bug that caused a timeout in the frequency filter and failed loading of the associated analysis.
# Version 1.11.2
Release date: 27.10.2020
# Highlights
This release adds bugfixes related to manually appending results to an analysis.
# All changes
- Fixed a bug where filters using
inheritance_modefailed to load when there are more than two proband samples in an analysis (e.g. on manual appending import). - Fixed a bug where manually appending an import to a finalized analysis fails.
# Version 1.11.1
Release date: 21.08.2020
# Highlights
This release adds a few bugfixes and improvements.
# All changes
- Implemented a workaround for an igv.js bug (opens new window) to properly distinguish reads with mapping quality 0 from other reads (these will be now be shown with lighter colors).
- Added support for haploid genotypes.
- Added CLI support for deleting allele interpretations and deleting analyses with associated allele assessments.
- Fixed bug causing incorrect calculation of class with ACMG criteria PS* + 2 PM*.
# Version 1.11
Release date: 25.06.2020
# Highlights
This release brings an improved gene information popup with editing possibilities, and several changes and improvements to the OVERVIEW page.
# User-editable gene information
The gene information popup (click the gene name in the top bar) has been improved, with the possibility to add and edit comments about the gene. This can be used for information that is important for evaluating variants in this gene, and is available for all analyses where the gene is included.
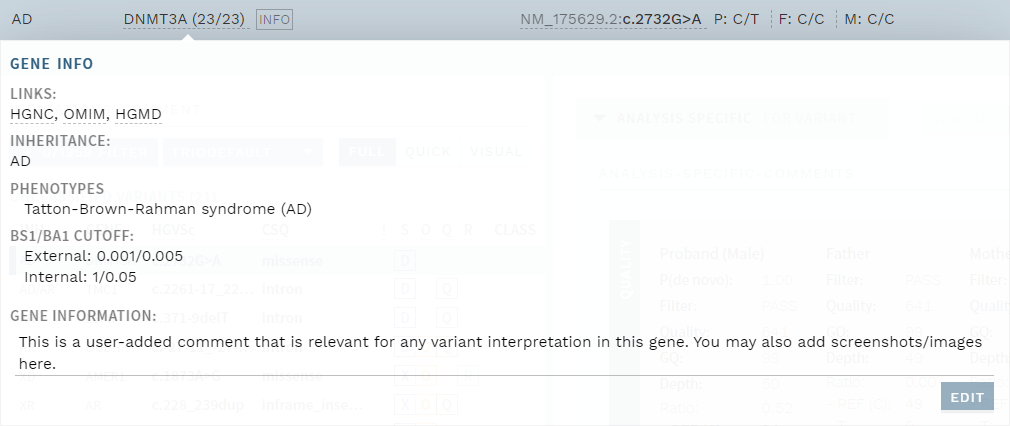
Figure: User-editable gene information.
If a comment has been added, an INFO tag is shown next to the gene name.
# New OVERVIEW filter feature
A new filter feature was added to the ANALYSES OVERVIEW page to quickly locate subsets of analyses. You can filter by these parameters:
- Analysis name
- Comment text (e.g. useful with auto-comments)
- Date requested (ranges up to current date)
- Show only
- HTS/Sanger
- Priorities Normal/High/Urgent
Any combination of these filters is allowed.
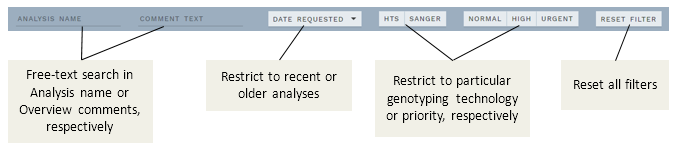
Figure: User-editable gene information.
Note that the filters do not include finalized analyses.
# Optional OVERVIEW sections replaced with auto-comments
This release retires the optional classification status sections on the OVERVIEW page and replaces them with a possibility to auto-add comments (ALL CLASSIFIED/NO VARIANTS) upon deposit of new analyses to the ELLA database. In addition, the VARIANTS OVERVIEW page has been limited to manually imported, stand-alone variants or individual variants opened from search.
# Option to set GQ thresholds for de novo candidates
To remove false positive de novo predictions in the segregation filter, it is now possible to set a genotype quality (GQ) threshold. This will disregard any de novo prediction where the proband, father or mother in a trio has a GQ value below the given thresholds.
# All changes
- Added possibility to edit and save gene information.
- Added filtering feature in the ANALYSES OVERVIEW.
- Replaced optional ANALYSES OVERVIEW sections with auto-comments
- Added option for GQ threshold for de novo candidates
- Fixed a bug causing previously cleared warning tags to remain in the Finalized section and in search results.
# Version 1.10.1
Release data: 15.06.2020
# Highlights
This release fixes a few bugs bugs related to references and reference assessments.
# All changes
- Fixed a bug where existing reference assessments would not show if no longer part of annotation
- Fixed a bug where ClinVar references would not be recognized during import of ClinVar annotation
- Fixed a bug where HGMD reference comments would display un-translated character sequences
# Version 1.10
Release date: 09.06.2020
# Highlights
This release brings improvements to the variant filters, a new version of IGV for Visual mode, as well as several smaller improvements and fixes.
# Improved variant filters
All criteria in the segregation filter can now be enabled/disabled separately, allowing for increased flexibility. In addition, the quality filter can now use the VCF FILTER status (PASS/...) as a parameter.
# All changes
- Added configuration for segregation filter.
- Renamed "Inherited mosaicism" to "Parental mosaicism"; The associated
Mtag can now appear together with other Segregation tags in the sidebar if multiple conditions are true. - Added possibility for using VCF
FILTERstatus in quality filter. - Removed
QUAL≤300 as a criterion for the NEEDS VERIFICATION warning and theQtag. - Upgraded IGV (in Visual mode) to version 2.5.4, with several minor improvements.
- The number shown on the
WORK LOGbutton now includes a count of all user added messages, including from previously finalized analysis rounds. - Removed
VARIANT REPORTbutton from OVERVIEW. - Open the previously selected overview page when returning from search.
- Fixed a bug causing wrong open-end position for insertions.
- Fixed a bug causing incorrect filtering for regions with 1 base.
- Fixed a bug causing incorrect rescue of variants annotated with non-standard terms in the ClinVar database.
- Fixed a bug causing a wide sidebar with long indication comments.
- Fixed a bug causing missing word wrap in comment fields.
# Version 1.9.2
Release date: 27.04.2020
# Highlights
This release contains a few bug fixes and improvements. Notably, support for the new PubMed page was improved, changing the recommended import procedure to use the new Save option to download the necessary text file (see the documentation for further details):
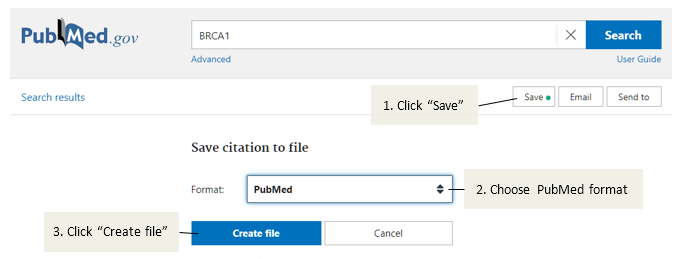
Figure: New recommended procedure for downloading PubMed references.
# All changes
- Added conversion of phased data to unphased data on import.
- Added VEP consequence
start_retained_variantto rules. - Added support for PubMed/MEDLINE format in reference import.
# Version 1.9.1
Release date: 26.03.2020
# Highlights
This version adds bugfixes and improvements to the finalize variant functionality introduced in version 1.9, support for a new version of VEP, as well as several minor UI improvements.
# All changes
# Finalize variant fixes and improvements
- Disallow finalize for an analysis when there are unsaved changes in REPORT (CLASSIFICATION page, user must push
SUBMIT REPORTfirst). - Added button for undoing changes to REPORT comment on CLASSIFICATION page.
- Fixed issue where variants present in multiple proband samples within an analysis could not be finalized.
# Support for new VEP version
Added support for updated version of VEP (v98.3), including fixes for:
- Fetching latest HGNC gene symbol.
- Choosing the correct RefSeq transcript version. If possible, the version specified in the gene panel is chosen, otherwise the latest available version is used.
# Other UI improvements and bugfixes
- Added auto-scroll to top in main window when switching between variants in ANALYSES view.
- Changed
HI FREQandHI COUNTin QUICK mode to display0instead of-when all sources report 0 frequency. - Adjusted position of modals to allow viewing variant information in top bar when modal is open.
- Removed
COPY ALL TO ALAMUTbutton. - Changed default view in
WORK LOGtoMESSAGES ONLY. - Increased custom gene panel name character limit to 20.
- Increased height of batch filter box when creating custom gene panels.
- Adjusted delay for popovers.
- Improved help text for RIS format import.
- Fixed word wrap in popover comment fields (ACMG comment and
WORK LOG). - Fixed an issue causing wrong page height and extra scrollbars.
- Fixed bug where variants with no transcripts were not filtered on frequency.
- Fixed an issue where "Worse consequence" warning would show if no consequence was available.
# Version 1.9
Release date: 31.01.2020
# Highlights
This version adds changes to better support larger number of users and user groups using the same variant interpretation database.
# Finalize variants in analyses
The most important change in this version is the addition of a "finalize variant" function in analyses. This allows variant interpretations to be released for overlapping analyses (where the same variant is present in more than one non-finalized analysis workflow) as quickly as possible. This also means that workflow collision warnings are given per variant instead of per analysis, reducing the number of displayed warnings.
With this change, each variant with a new/updated classification in an analysis needs to be finalized by pressing the new FINALIZE button in the CLASSIFICATION section before the analysis (sample) itself can be finalized:
Figure: New button to finalize variant in analysis.
Note that this also needs to be performed for any classification done in QUICK mode, and that interpretations done in VARIANTS mode (stand-alone variants e.g. from search) still need to be finalized as before via the FINISH button in the top bar before the variant is considered truly finalized.
# User group warnings
Finalizing variants means each variant interpretation will be tagged with the main responsible user and user group. This also allows showing a warning if the previous (finalized) variant interpretation was performed by a user from another user group than your own:

Figure: New user group warning.
# Copy text by mouse-click
You can now more easily copy text from many pop-overs by clicking on the text. Look for the clipboard symbol, e.g. in the HGVS cDNA variant name in the top bar:
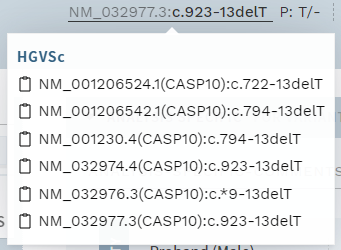
Figure: New option to copy text directly by mouse-click in popups.
# All changes
- Finalize variant in analysis
- Display warning when variant was finalized by different group
- Mouse popovers and tooltips now use tippy.js (opens new window), with possibility for copying text by mouse-click (any text marked with a clipboard symbol).
- Optional sections on the ANALYSES OVERVIEW page have been simplified, see ANALYSIS worklist.
- Added analyses with variants that need verification for optional automatic sorting to NOT READY section on the ANALYSES OVERVIEW page (see Deposit).
- Added support for importing references with .RIS format from the new PubMed webpage (opens new window).
- Entries listed in the STUDIES & REFERENCES section are now shown with annotation source and better separation.
MARK CLASS 2button has been removed from the FREQUENCY section (CLASSIFICATION page).- Added condition to the Inherited mosaicism filter to exclude cases where the other parent has a normal genotype.
- ACMG criteria now have short descriptions available everywhere.
- All action buttons are now disabled until all data has been loaded when opening an analysis.
- Improve the performance of the region filter
- Most configuration settings that were previously hard coded in
/src/api/config/config.pyhave now been replaced by a dynamic configuration, set by the environment variableELLA_CONFIG. See/example_config.ymlfor examples. - Fixed issue during import when re-importing previously deleted analyses.
- Changes to the documentation, including moving the "Concepts" section to the User manual.
# Version 1.8.2
Release date: 02.12.2019
# All changes
- Fixed bug where quality filtering did not work for multiple probands.
- Fixed bug where importing the same sample more than once caused a crash.
- Fixed bug causing several variants not to be filtered out as they should in the region filter.
# Version 1.8.1
Release date: 30.10.2019
# All changes
- Fixed bug where users were not able to finalize analyses with reused allele assessments and auto-ignored references under "Pending evaluation".
- Fixed bug where reference data was not reloaded when including a variant from filtered variants.
# Version 1.8
Release date: 30.10.2019
# Highlights
# Add "other" and unweighted ACMG criteria
Sometimes, criteria that don't match the ACMG guidelines are important for a variant interpretation, e.g. the ENIGMA criteria for the BRCA1/BRCA2 genes. ELLA now allows adding these to the interpretation as a generic OTHER criterion. The type and impact on the classification should be given in this criterion's comment field once added.
In addition, users can often spend significant time evaluating an ACMG criterion for a particular interpretation, but in the end decide that the requirements are not met. ELLA now allows setting an added ACMG criterion as NOT WEIGHTED, so that comments related to this work can be properly recorded.
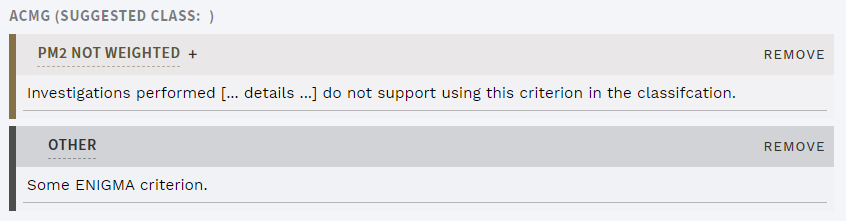
Figure: The new "other" criterion and unweighted option for ACMG criteria.
Note that neither "other" or unweighted ACMG criteria are counted in the calculation of suggested classification.
# Filter improvements: Gene and allele ratio
The filter settings now allows using genes as a variable in rules for filters or exceptions. This allows conditioning any rule on the presence/absence of a gene, e.g. exclude certain genes from a particular filter.
In addition, it is now also possible to use allele ratio (alternative allele reads/total reads) as a variable in the quality filter. In our experience, this gives a more powerful filter than using the qual variable, especially for high sequencing depths. A caveat is that mosaic variants may be missed, but this can be partially be circumvented by adding particular genes where mosaics are expected to a gene exclusion rule as described above.
# All changes
- Added possibility for adding non-ACMG criteria.
- Added possibility for setting ACMG criteria as unweighted.
- Added possibility to filter out or rescue variants in specific genes.
- Added possibility to use allele ratio in quality filter.
- Added possibility to configure irrelevant references to be automatically IGNORED; see Technical documentation for details.
- Made controls FULL - QUICK - VISUAL and INDICATION comment field in the sidebar sticky to reduce need for scrolling when there are many variants.
- Made collision warnings below the top bar sticky and collapsible.
- Increased number of retrieved reference search results.
- Added "Documentation" link to all pages.
- Added possibility to copy gene + transcripts from filtered results in GENE PANEL INFO.
- Fixed bug that caused non-working filtering in GENE PANEL INFO if using wrong case.
- Fixed bug resulting in empty interpretation window when no variant was selected.
- Fixed bug causing no alerts when navigating away from interpretation view with unsaved work.
- Import jobs now show time of day when job was started, and time stamp when status of job last changed.
- Improved Technical documentation.
# Version 1.7
Release date: 05.09.2019
# Highlights
# Improved import
Import of variants in various text formats and ordering of reanalyses from existing samples is now merged into a single import section on the OVERVIEW page, replacing the old IMPORT button in the top bar.
The new import solution includes the possibility to adjust the priority (for the OVERVIEW work lists) for any import involving a sample. In addition, it is now possible to search for multiple genes at once (batch query) when ordering a custom reanalysis, e.g. by pasting a list of genes from an external source. Both gene names and HGNC ID is supported, and any genes not found are shown as a list that can be copied out.
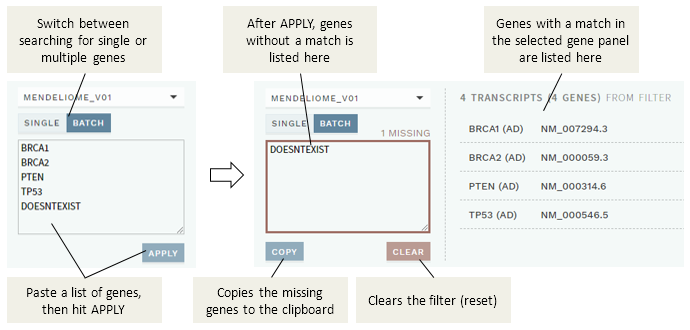
Figure: The new batch filter mode for searching for multiple genes when ordering a custom reanalysis.
# Gene panel info
Various information about the gene panel used in an analysis is now available via a new button in the top bar:
The information includes which genes are in the panel (with inheritance and default transcript available on mouse-over; list can be copied), as well as a list of the five most similar gene panels:
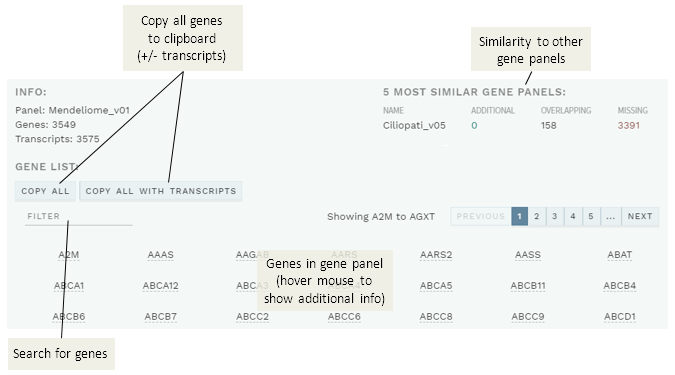
Figure: Gene panel info.
# Improvements to attachments
Attachments are now named after the filename (instead of an index number), and details are available by hovering the mouse over an attachment.
# All changes
- Merged import functions into single section on OVERVIEW page.
- Added possibility for setting priority on manual imports and reanalyses.
- Added possibility for batch queries of genes to include in a reanalysis.
- Added gene panel info.
- Attachments named after filename, mouse-over gives details.
- Harmonized formatting of REPORT field on CLASSIFICATION and REPORT pages.
- Order of alleles is now consistently REF-ALT in the QUALITY card.
- Fixed bug causing missing REPORT comment field for certain, remnant HTML formatting.
- Fixed bug causing timeouts when changing workflow state for analyses with large gene panels.
# Version 1.6.2
Release date: 09.08.2019
# Fixes
- Fixed performance bug that caused excessive loading time for ANALYSES overview.
# Version 1.6.1
Release date: 27.06.2019
# Fixes
- Fixed add/remove buttons for variants not working in REPORT side bar.
# Version 1.6
Release date: 16.06.2019
# Highlights
This update brings a few improvements to the user interface and workflow, in addition to several bug fixes.
# Quick check boxes for BS1/BS2 and ACMG indicators
The most noticeable changes are the addition of selection buttons for ACMG criteria BS1 and BS2 (and shortened button titles) in the QUICK CLASSIFICATION mode, and addition of indicators for added ACMG criteria in the side bar.
Note that to select CLASS 2 for a variant in QUICK CLASSIFICATION mode, you must now first add at least one benign ACMG criterion (e.g. BS1/BS2 via the check boxes); see the figure below.
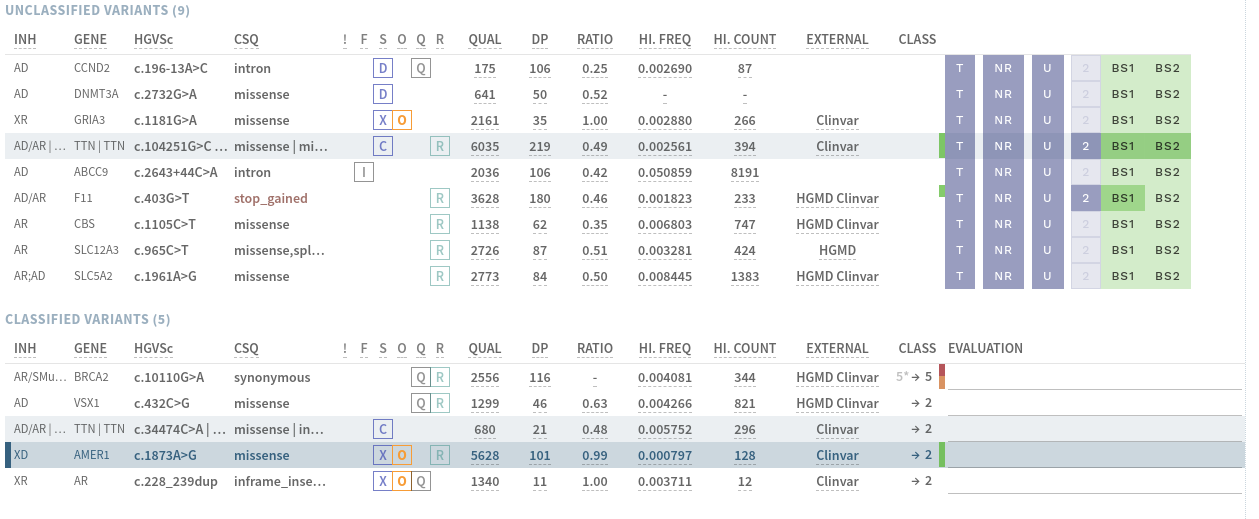
Figure: Check boxes for BS1 and BS2 for CLASS 2, and ACMG indicators (vertical bars next to CLASS column) in side bar.
# Outdated variants treated as UNCLASSIFIED
This update also brings a few changes to how outdated variant interpretations are handled. These are now grouped with UNCLASSIFIED variants in the side bar of an analysis, and must be reopened individually by clicking the RE-EVALUATE button if a new evaluation is to be made. This is to ensure a new validity period is only set for variants that have actually been re-evaluated.
As an added measure, the classification of reopened variants must also be actively reselected.
# All changes
- QUICK CLASSIFICATION mode: Added check boxes for ACMG criteria BS1 and BS2 (for class 2), shortened button titles.
- Side bar: Added indicators for added ACMG criteria (full list on mouse-over).
- Disallow adding the same ACMG criterion (irrespective of strength modifications) more than once to the same variant interpretation.
- Sort added ACMG criteria by pathogenic-benign, then strength.
- Variants with outdated interpretations are now grouped with UNCLASSIFIED VARIANTS in the side bar in an analysis, and are no longer automatically reopened with the analysis.
- Added possibility to edit comments on individual studies in STUDIES & REFERENCES section directly in the list (without opening the evaluation form) after first evaluation.
- When there are multiple VEP CSQs (consequences) for a variant, these are now sorted in the side bar by severity (worst consequence first).
- TECHNICAL button is now available also for CLASSIFIED VARIANTS in VISUAL.
- Added subtitle "FOR VARIANT" to the ANALYSIS SPECIFIC section on the CLASSIFICATION page, to avoid confusion (some users mistook the section to apply to the analysis as a whole, not the particular variant).
- REPORT page: Added homo-/hemizygous variants were not correctly HGVSc-formatted. As a temporary fix (until formatting is properly fixed), all HGVSc is now formatted with empty brackets for nucleotide changes, to be filled out by the user.
- Fixed bug where variants from other user group showed up in the VARIANTS overview.
- Reference evaluation form: Renamed
SAVEbutton toCLOSE, removedCANCELbutton. - Fixed missing mouse-over for
Mtag in the side bar.
# Version 1.5
Release date: 23.05.2019
# Highlights
# Classification view modes
The VISUALIZATION page has been redefined as a view mode on the CLASSIFICATION page, allowing for easier switching between relevant views while performing variant classification. Switching between view modes can now be done with three buttons above the variant list in the side bar:
FULL(new button; default view mode).QUICK(renamed from QUICK CLASSIFICATION).VISUAL(renamed from VISUALIZATION; button moved from top bar).
# Changes to side bar functionality
It is now possible to mark variants as TECHNICAL in the side bar in VISUAL mode (with commenting after marking, similar to QUICK mode), and the INDICATIONS COMMENT from the REPORT page is mirrored in the side bar on the CLASSIFICATION page, allowing for a more efficient workflow.
# ACMG modified criteria according to ClinGen
ACMG criteria where the strength of the original criterion are now displayed according to ClinGen's recommendations, e.g. PM1_Strong instead of PSxPM1.
# All changes
- VISUALIZATION redefined as view mode VISUAL on CLASSIFICATION page, with navigation moved to the side bar.
- Added function to mark as
TECHNICAL(button and comment) in VISUAL mode. - Mirrored INDICATIONS COMMENT from the REPORT in the side bar on the CLASSIFICATION page.
- Changed naming of ACMG criteria with modified strength to ClinGen's recommendations.
- Added QUAL column to variant lists in the side bar for analysis workflow (QUICK and VISUAL mode) and filtered variants.
- Adding/removing variants in the REPORT restricted to toggle button (not clicking the anywhere on the variant).
- Fixed sorting on F column (tag: Included) in the side bar.
- Fix issue where gene panel was not reloaded when including a filtered variant.
# Version 1.4.1
Release date: 15.04.2019
# Additions and fixes
- Increase overview update interval to lessen strain server.
- Fix automatic import of analyses with underscore in the gene panel name.
- Fix issue where finalization would not work under certain conditions.
# Version 1.4
Release date: 26.03.2019
# Highlights
# Variant filter configurations
ELLA now supports configuring several filter configurations for a user group. This lets you define filter chains that are specific to certain types of analyses (e.g. single or trio analyses) or specific analysis names. One analysis can have several applicable filter configurations which will show up as options in the side bar inside the analysis workflow:
Figure: Switch between different filter configurations in an ongoing analysis.
Examples of possible filter configurations are different levels of frequency thresholds, or being able to turn on and off family (segregation) filtering for an analysis.
# New variant filters
Current filters include:
- Classification filter
- Consequence filter
- External filter
- Frequency filter
- Inheritance model filter
- Polypyrimidine filter
- Quality filter
- Region filter
- Segregation filter
See new section Concepts/Filters in the documentation for details on how these filters work and how to configure them.
# Comment editor additions
The comment editor now supports inserting pre-defined templates, as well as references from the STUDIES & REFERENCES section, using two new buttons in the comment field menu. Templates can be defined independently for different comment fields, and supports basic text formatting.
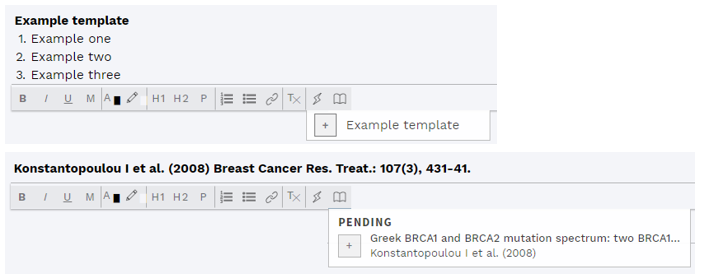
Figure: Adding templates and references.
If no template has been defined or no reference has been found/added (STUDIES & REFERENCES section), the respective buttons will be inactive (greyed out).
# New features
- Variant filter configurations.
- New filters (turn on/off in configuration).
- Use text templates and add references in comment fields.
- Work log: Hide system messages by clicking
MESSAGES ONLY. - New class: "Drug response" (as defined in ClinVar (opens new window)).
- New "Indication comment" on the REPORT page, for comments specifically about the patient's indication.
- Nearby variants warning now checks all variants in analysis (previously only checked non-filtered variants). Now also checks nearby within 2bp (possibly same codon), rather than 3bp.
- Empty gnomAD, ClinVar and HGMD Pro records now have external links to generic pages.
- Added warning for expiring passwords.
# Other additions and fixes
- Added
ADD STUDIESbutton in top bar. - Fixed description for ACMG criterion BP6.
- Fixed missing scrolling in "Show analyses".
- "PL" removed from Quality card in ANALYSIS SPECIFIC section.
- Changed order of sections on CLASSIFICATION page to PREDICTION - EXTERNAL - STUDIES & REFERENCES
- References are now ordered (descending) according to publication year.
- Changed colour of
Otag in side bar.
# Backend
- Added broadcast functionality to convey important messages in ELLA to all users.
- Improvements to filter efficiency and speed.
# Version 1.3.1
Release date: 21.12.2018
# Additions and fixes
- Fix issue in
Variant reportwhen parsing warning from pipeline, in some cases yielding wrong number of poorly covered regions.
# Version 1.3
Release date: 14.11.2018
# Highlights
# Visualization (analysis workflow)
As part of this release, igv.js (opens new window) has been integrated into ELLA as part of a new visualization feature. ELLA now let's you visualize all variants in an analysis, along with user customizable tracks at three different levels: global (all users), user group and analysis.
ELLA provides a few special tracks by default:
- Gene panel: Shows the analysis' gene panel.
- Classifications: Shows all classifications present in the database.
- Variants: Shows the analysis variants after filtering.
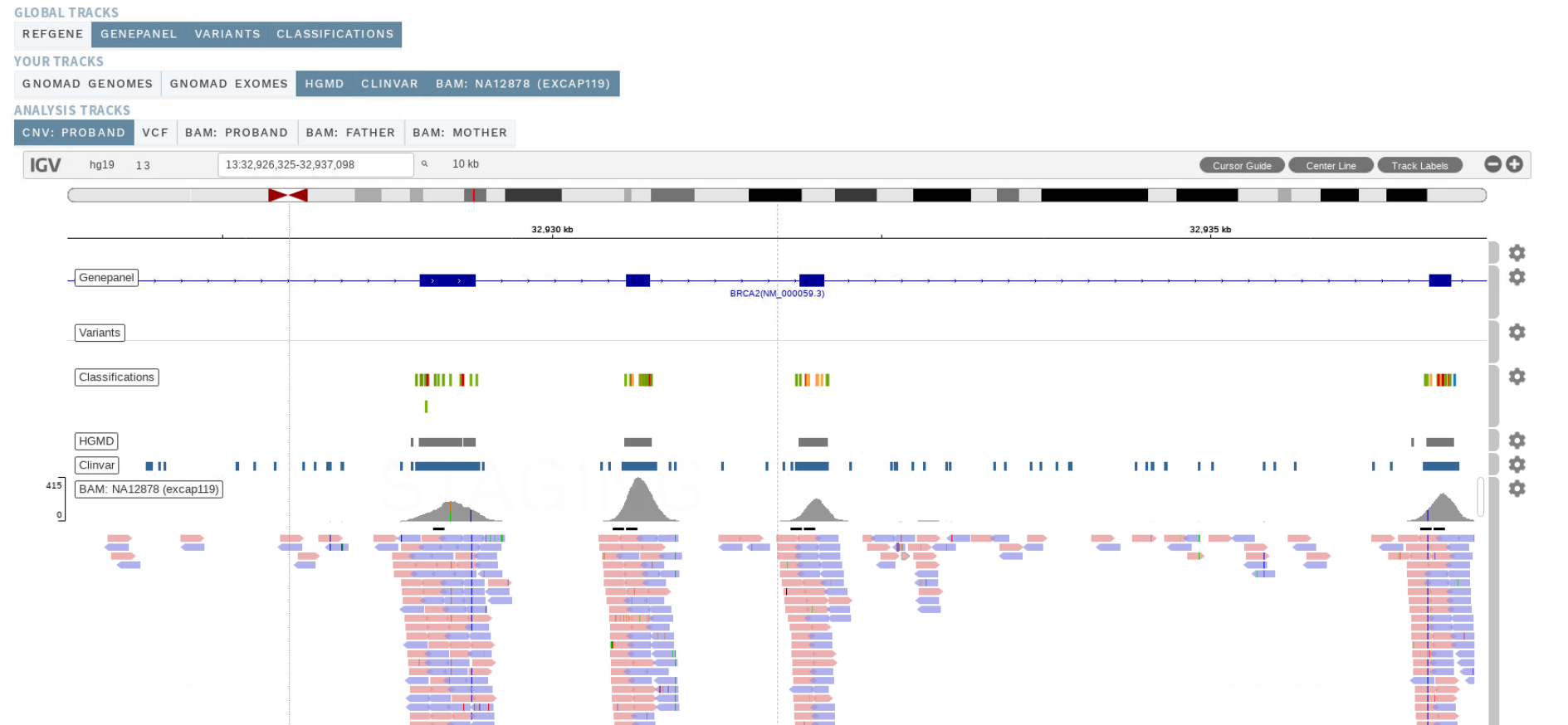
Figure: New visualization feature.
# Mark "Not relevant" (analysis workflow)
Variants can now be marked as Not relevant for the analysis. Such variants can, depending on the user's configuration, be left without a selected class upon finalization of the analysis.
# Quick classification (analysis workflow)
A new quick classification view is now available, aimed at certain workflows for large analyses with many variants.
It gives a summary of the most important information necessary for classifying variants as Technical, Not relevant or Class 2.
It is most relevant for workflows where you first perform a quicker interpretation of certain variants, before doing a more thorough interpretation of the remaining ones.
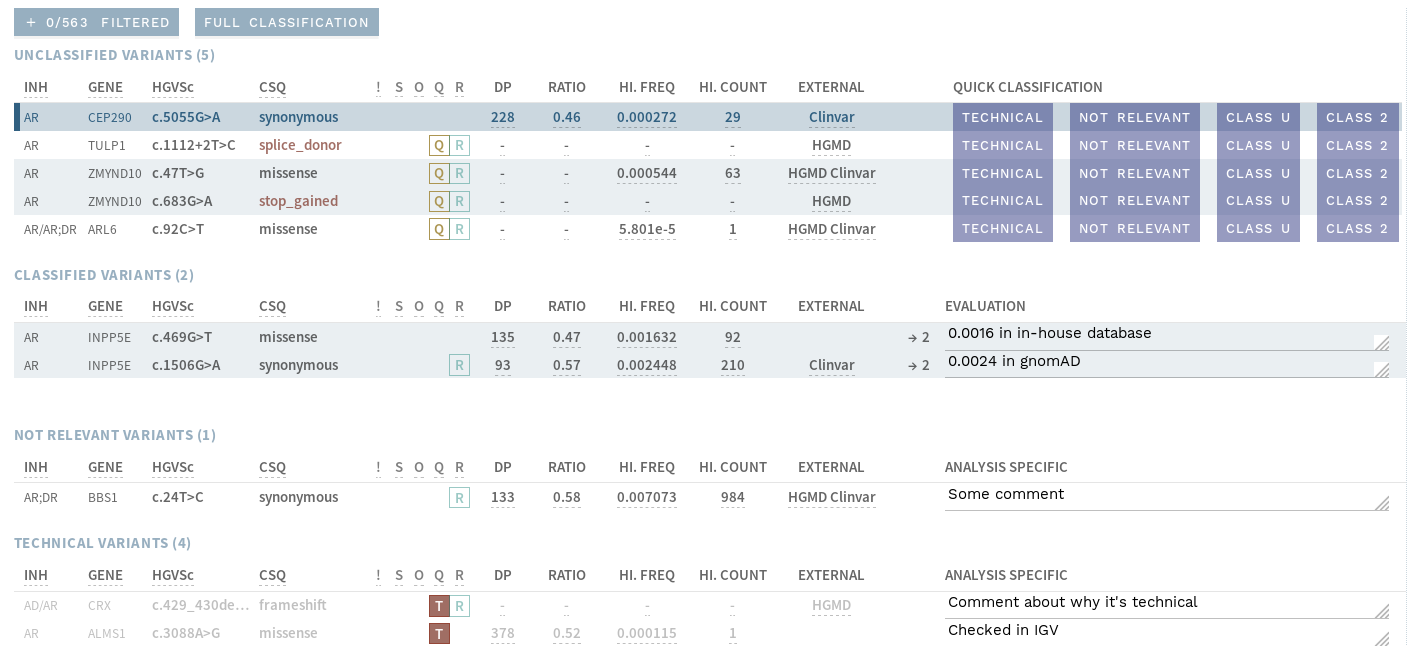
Figure: Quick classification view.
# QUALITY card renamed to ANALYSIS SPECIFIC (analysis workflow)
The card in Classification view previously referred to as QUALITY is now called ANALYSIS SPECIFIC to highlight the fact that it's not part of a variant's classification, but rather just a part of the analysis.
The card is blue and collapsed by default, to further separate it from the classification related cards.
# Improved view of existing and current class in sidebar (analysis workflow)
The view of a variant's class in the sidebar has been improved.
Left number is existing class, right is new. An arrow indicates that a new classification will be created. Blue background indicates that the variant has been reviewed. You can toggle the review status by clicking on the class in the sidebar.
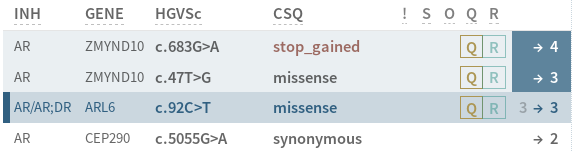
# New user manual
A new, online user manual is now available from within ELLA itself. You can access it by clicking Documentation in the top navigation bar in the overview.
# New features
- Visualization (analysis workflow).
- Ability to mark variants
Not relevant(analysis workflow). - Quick classification (analysis workflow).
- Filters and their parameters are now configurable per user group
- New filter: Quality
- Region filter now can save variants with certain (configurable) consequences from being filtered.
- New variant warning: HGVSc and HGVSp mismatch between corresponding Refseq and Ensembl transcripts.
- Integrated documentation within ELLA.
# Other additions and fixes
- Filtered variants are now shown as a list (analysis workflow).
QUALITYcard is renamed toANALYSIS SPECIFIC(analysis workflow).- Improved view of class in sidebar (analysis workflow).
- Workflows can now be finalized with technical, not relevant and/or missing classifications (depending on configuration). Workflows can still force valid classifications for all variants if desired (old behavior). (analysis workflow)
Requested dateis now read from input.analysisfile and used in overview.- Too wide images in comments will not make the page scrollable in the horizontal direction.
- Overview comment is now visible for
Finalizedanalyses and variants in overview. - Low quality warning is removed for Sanger variants, as there is no quality data.
- HTML content is now properly sanitized when pasted into comment fields.
- Fix issue where technical status was not reflected in the
TECHNICALbutton in theQUALITYcard under certain conditions. - Fix issue where image could not be resized in reference evaluation.
# Version 1.2
Release date: 02.10.2018
# Highlights
# Family analysis
ELLA now lets you interpret analyses with variants that have been joint called within a single family.
The following segregation models are supported:
- De novo
- Autosomal recessive homozygous
- X-linked recessive
- Compound heterozygous
The most powerful filtering requires two parents to be present in the analysis, but some segregation filters and tags also apply to analyses with only siblings (unaffected and/or affected).
Figure: Segregation tags in sidebar.
Variants filtered out by these filters can be found in the Segregation category in the excluded variants.
# Work log
The analysis and variant workflows now have their own Work log. The work log currently lets you:
- Update the
Overview comment(previouslyReview comment) - Clear any analysis warnings (analysis only). Clearing a warning makes the warning tag disappear from the Overview.
- Adjust the priority of the analysis or variant
- Add messages that should be available to yourself or later interpreters about things that are relevant for the interpretation of the analysis or variant. Messages can contain formatting and images, and are editable until the next interpretation round is started.
All options in the work log can be changed at any time, without having to start a new interpretation round.
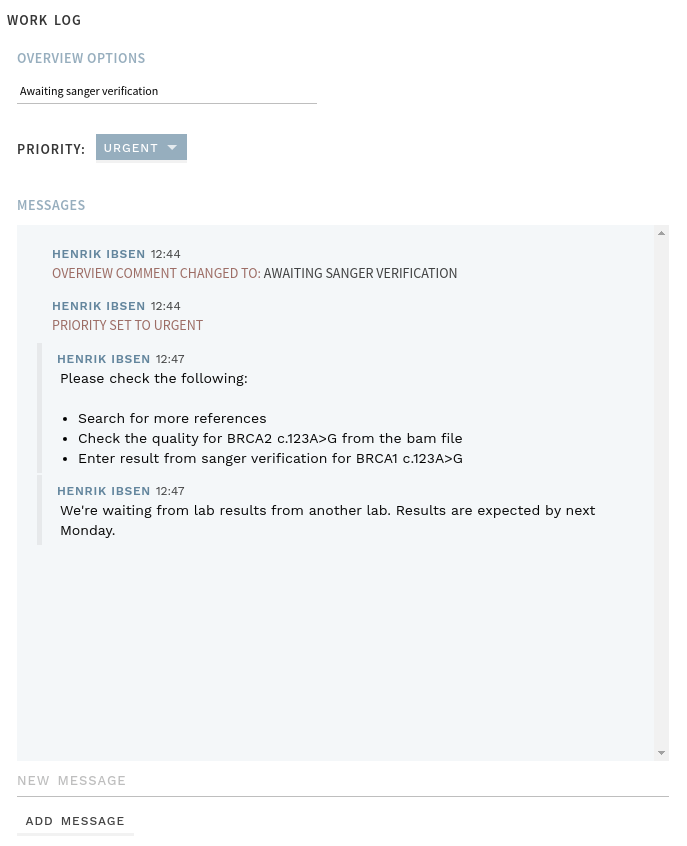
Figure: Work log example.
If there are any messages since last time the workflow was Finalized, the work log button will appear in purple, along with the current message count.
Figure: 2 messages since beginning or last finalization.
# Variant warnings
Variants are now tagged with warnings whenever there is something special that considered for the variant in question. The list of warnings will be expanded later, but currently includes:
- Worse consequences in other transcripts
- Other variants are within 3 bp of the variant in the analysis
Variant warnings are implemented for both the variant and analysis workflows, but some warnings are only available for analyses.

Figure: Example warning.
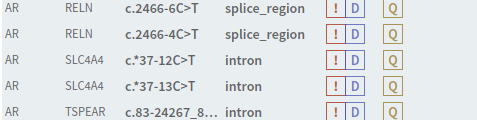
Figure: Warning tags in sidebar.
# New features
# Other additions and fixes
Qualityis now it's own section in Classification view (analysis only)- Quality verification for variants in an analysis (
VerifiedandTechnical) is moved from the Info view to the Quality section in the Classification view. - Variants marked as
Technicalare moved to it's own list in the sidebar. - Improvements in display of variants with multiple selected transcripts.
# Version 1.1.2
Release date: 18.07.2018
# Additions and fixes
- Add red 'HOM' tag to top variant bar in order to improve homozygous visibility.
- Merge
utrandintronfilters into a new, improvedregionfilter. - Improve search performance (entering a gene is now required for searches using HGVS nomenclature).
- Show more information about the available samples in import view
- Add new external database for gene TP53
- Fix missing Hemi total count for gnomAD
# Version 1.1.1
Release date: 24.05.2018
# Additions and fixes
- Add BRCA Exchange to external databases for BRCA1 and BRCA2
- Add ability to search using genomic position on format g.123456
- Keep existing reference evaluation data when clicking 'Ignore'
- Fix link and reference description in reference evaluation window.
- Fix issue where some variants would appear with two gene panels in variants overview
- Fix issue where worst consequence would not display correctly for a rare case with variant having intron_variant as consequence in one transcript and splice_region_variant, intron_variant as consequences in another transcript.
- Fix issue importing Pubmed XML data for some references.
# Version 1.1
Release date: 15.05.2018
# Highlights
# New import functionality
Requires access to the import view.
ELLA now lets you re-import previously run samples, using either an existing gene panel or a gene panel customized for that specific sample.
This lets you request new analyses directly in the application and shortens the time for reanalysis with a different set of genes.
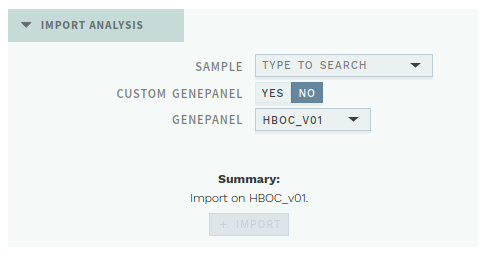
# Frontend code improvements
The frontend code has been refactored to make it more responsive and to make it easier to add new functionality going forward.
# Other additions and fixes
- Display number of excluded references on 'SHOW EXCLUDED' button
- Remove scrollbar on comment fields.
- When there are multiple transcripts in a gene panel, sort them by name. Also display all transcripts in more places, for example in the variants overview.
- Do not add references with Relevance: 'No' to the excluded references list.
- The 'ADD EXCLUDED' window for adding excluded variants now loads faster.
- Search results will now show correctly when typing quickly.
- Many other smaller UI fixes