# Visual mode
Pushing the VISUAL button (available in ANALYSIS mode only) in the side bar opens the VISUAL mode:
This mode features an integrated version (opens new window) of Integrative Genomics Viewer (IGV) (opens new window):
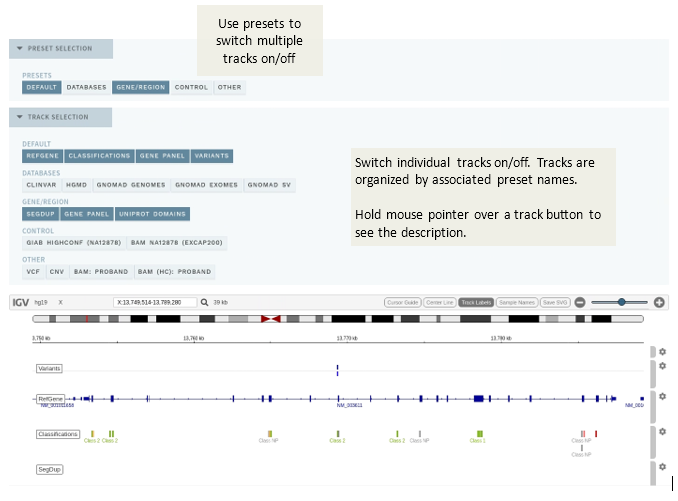
# Preset selection
This section lists all presets defined for individual tracks and allows you to quickly switch on/off all tracks that are associated with the preset. The DEFAULT preset and associated tracks are turned on by default, while others must be turned on manually. See the technical documentation for details on configuration.
# Track selection
This section is collapsed by default. To switch on/off individual tracks, expand the section and click the associated track name. Tracks are grouped by their associated preset names.
To see a description of the track, hold the mouse pointer over the associated track button.
# Dynamic tracks
Some tracks are dynamic and generated on-the-fly in ELLA. These currently include:
VARIANTS: All variants in the same sample (analysis) after filtering.CLASSIFICATIONS: All existing (finalized) classifications present in the database. Clicking on a variant in this track gives a link to the associated allele assessment.GENEPANEL: Regions covered by the current gene panel.
Note: these tracks will usually be part of the DEFAULT preset (depending on configuration).
# Navigating the view
Options when navigating the view in VISUAL:
- Zooming: Press and hold the
ShiftorAltkey and use the mouse wheel, or the +/- buttons for more fine-grained control. - Panning: Click and hold the mouse button anywhere in the alignment and drag left or right.
- Recenter: You can quickly recenter on a variant (after panning) by selecting it again. Note that this also resets the zoom level to the default.
# Side bar: Mark as TECHNICAL
A button T is available in the side bar next to each variant for quickly marking technical variants. This works the same as in Quick classification mode, where clicking the button moves the variant to the TECHNICAL VARIANTS section in the side bar. If applicable, add a comment before moving to the next variant; this will also be reflected in the ANALYSIS SPECIFIC comment field on the CLASSIFICATION page.