# Side bar: Variant list, sorting and tags
The variants in your sample (ANALYSES mode only) are listed in the side bar to the left:
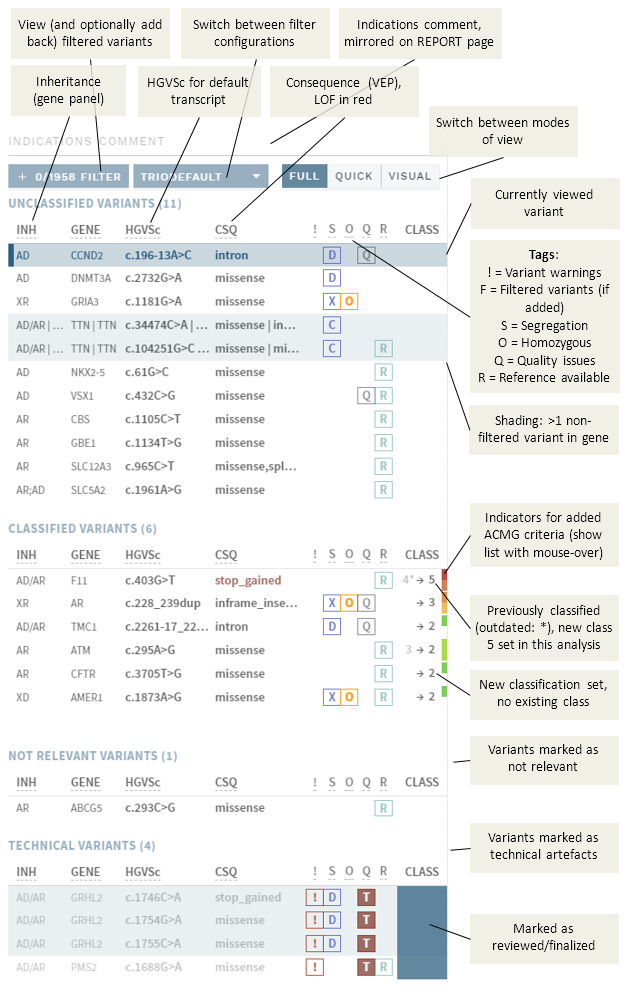
The side bar layout described here is for the default, full classification mode. The layout is almost the same in other CLASSIFICATION view modes (QUICK and VISUAL) and on the REPORT page, except for some columns/buttons. For details, go the referenced pages.
Note that names (variant HGVSc and gene) and inheritance are displayed as specified by the default transcript in the gene panel. If there is more than one default transcript, all versions are shown.
# Buttons and comment fields
The side bar has an INDICATIONS COMMENT field at the top, corresponding to the same field on the REPORT page (changes in either place is mirrored in both).
Directly beneath that are several buttons:
FILTER: View and optionally add back filtered variants.- [Drop-down menu]: Switch between (pre-defined) filter configurations.
FULL: Switch to Full (default; detailed) classification. See also Evidence sections, Classification section.QUICK: Switch to Quick classification mode.VISUAL: Switch to Visual mode.
# Side bar sections
Variants are listed in one of four side bar sections depending on the state of the variant interpretation:
UNCLASSIFIED VARIANTS: New variants with no existing interpretations (historic or current), or variants with previous classifications that are outdated and due for re-evaluation.
CLASSIFIED VARIANTS: Variants with previous interpretations that are still valid, or that have been given a new/updated classification in the current analysis.
NOT RELEVANT VARIANTS: Variants marked as not relevant for the current analysis.
TECHNICAL VARIANTS: Variants marked as technical artifacts in the current analysis.
# Variant sorting
The default sorting in each section is (Class) - Inheritance – Gene – HGVSc. You can change the sorting by clicking on any of the list headers (also in Quick classification):
- First click sorts descending (↓)
- Second click sorts ascending (↑)
- Third click returns to default sort
# Variant tags
If applicable, variants in the variant list are tagged with:
SSegregation. Depending on data, changes to:DDe novoAAutosomal recessive homozygousXX-linked recessiveCCompound heterozygousMParental mosaicism (may also occur in combination with other Segregation tags)
OHomozygous/hemizygous genotype.QQuality issues. Uses the same criteria that trigger the NEEDS VERIFICATION warning, except that insertions/deletions are not marked unless there are other quality issues. Depending on actions in the QUALITY section, this tag may be replaced by:VVerified (green)TTechnical (red)
RReference available (from annotation).IIncluded variants (columnFfor filtered, shown only if variants have been included from filtered variants).Shaded background: More than one variant in the same gene (in current sample).
NOTE
Presence of the M tag (parental mosaicism) often indicate quality issues rather than a parent actually being mosaic. E.g. MD indicates some uncertainty of the de novo status (D), as one or both parents have some evidence for the alternative allele (but not enough reads to be called heterozygous).
TIP
Hold the mouse cursor over a tag to see the full label.
# Existing and new classifications
Existing or newly set classes are given in the right-most column:

An arrow (→) indicates that a new classification will be created, and any existing class is given to the left of the arrow. If this classification is also outdated (long since last interpretation), an * is added to the existing class.
NOTE
Outdated variants marked with * in CLASSIFIED VARIANTS should be re-interpreted before you finish the analysis.
# Mark as reviewed/finalized
You can toggle a blue colouring of the background of the class by clicking on it in the side bar:

This is done automatically when you finalize a variant. Manual marking can be used to indicates which variants have been reviewed, i.e. to keep track of the work done in a review round.
# Experimental: CNV mode
NOTE
This functionality is currently disabled by default.
If the experimental CNV mode is enabled, selecting the CNV button in the top bar switches the view in the side bar to showing CNVs:
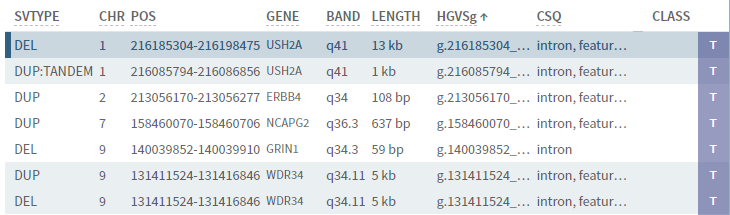
Except for the columns shown, sections and sorting works the same way as for SNVs.