# Filtered variants
Many variants have usually been automatically filtered from view before you start an analysis, based on pre-configured rules. These variants are still accessible via a button at the top of the variant list in the side bar:
In this particular sample, there are a total of 123 filtered variants, and one of these have been added manually to the analysis by the user.
Pushing the button brings up a window where you may select individual variants and add them to the analysis:
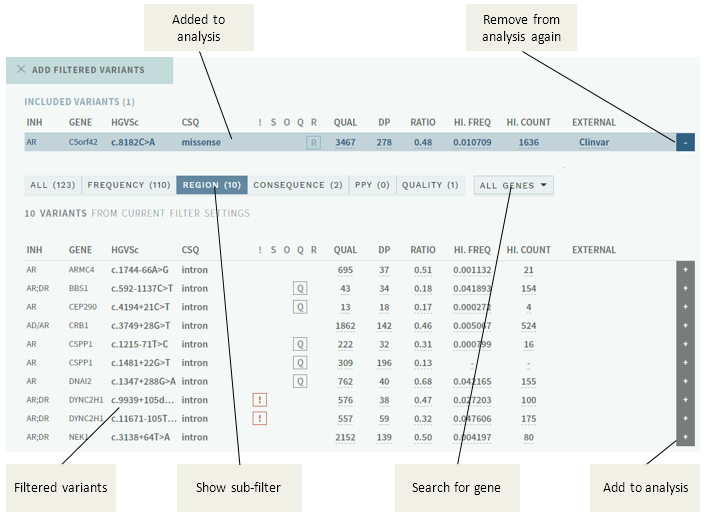
Pushing the + to the right of a variant adds the variant to the analysis, and will be shown in the side bar variant list with the I tag.
The filter categories ("sub-filters" in the figure above) are defined in the configuration for your user group/gene panel, and each may consist of a combination of rules for filtering and "rescue". The left-most category (ALL) shows all filtered variants combined, whereas the other categories are applied in the left-right order displayed (i.e. in the example above, variants are filtered on frequency first, remaining are filtered by region, etc.).
# Filter rules
Most filter rules may be configured to filter or "rescue" (exclude from filtering) variants, or a combination of both. See Technical documentation for further details and how to configure the filters.
Current rules include and use:
| Label | Based on |
|---|---|
| CLASSIFICATION | Existing classication in in-house database. |
| CONSEQUENCE | Predicted consequences from VEP (opens new window). |
| EXTERNAL | Reports/tags in external databases. |
| FREQUENCY | Population frequency above a threshold, with options for minimum allele number, gene inheritance model and dataset groups. |
| INHERITANCE MODEL | Other variants/alleles in same gene in single samples, depending on inheritance model for gene. |
| POLYPYRIMIDINE | Variants in the intronic polypyrimidine tract. |
| QUALITY | Quality criteria not met. |
| REGION | Outside a defined UTR/intron region. |
| SEGREGATION | Segregation patterns in family data. |